empower research, enrich diversity
Co-limitation towards lower latitudes shapes global forest diversity gradients
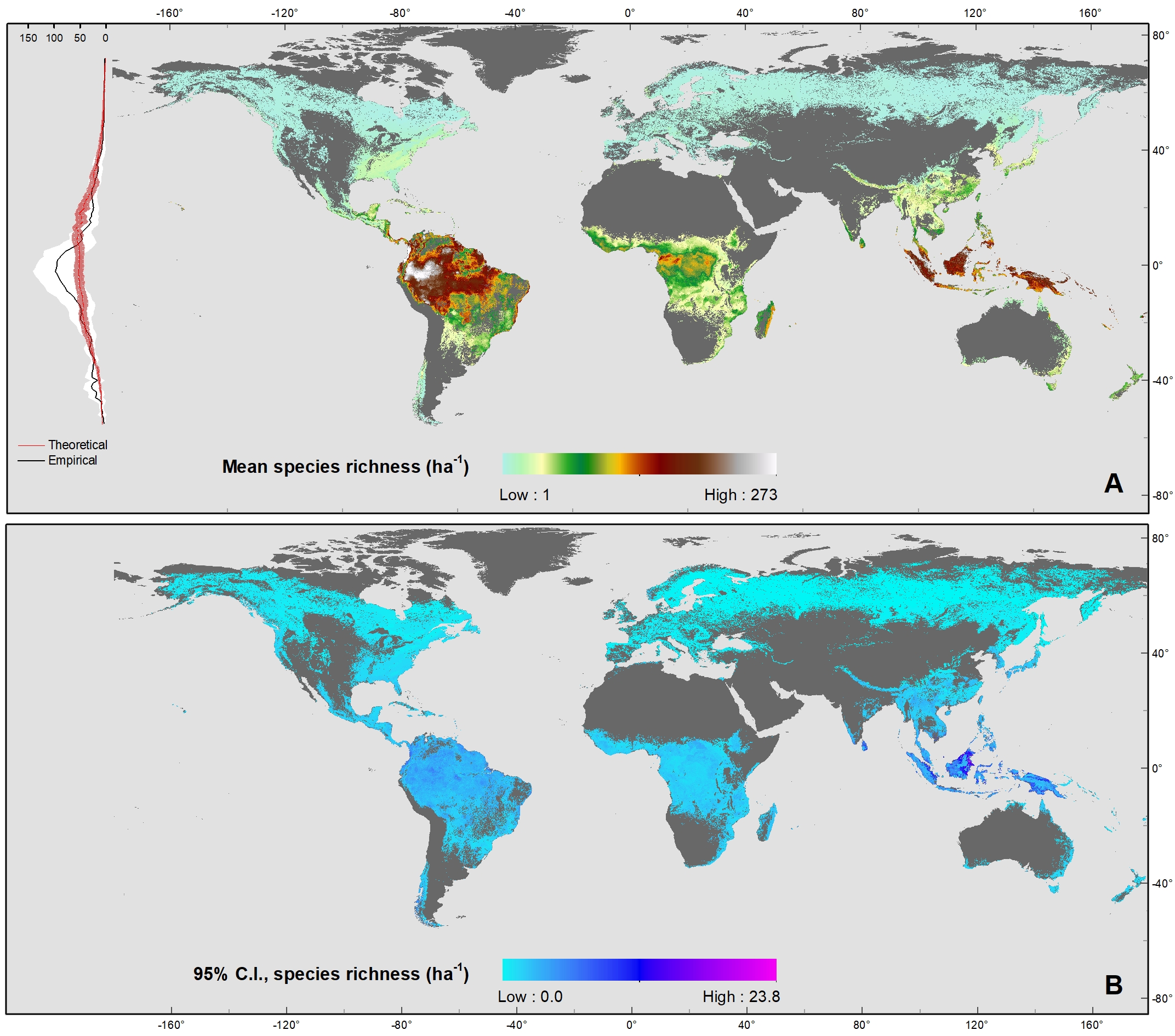
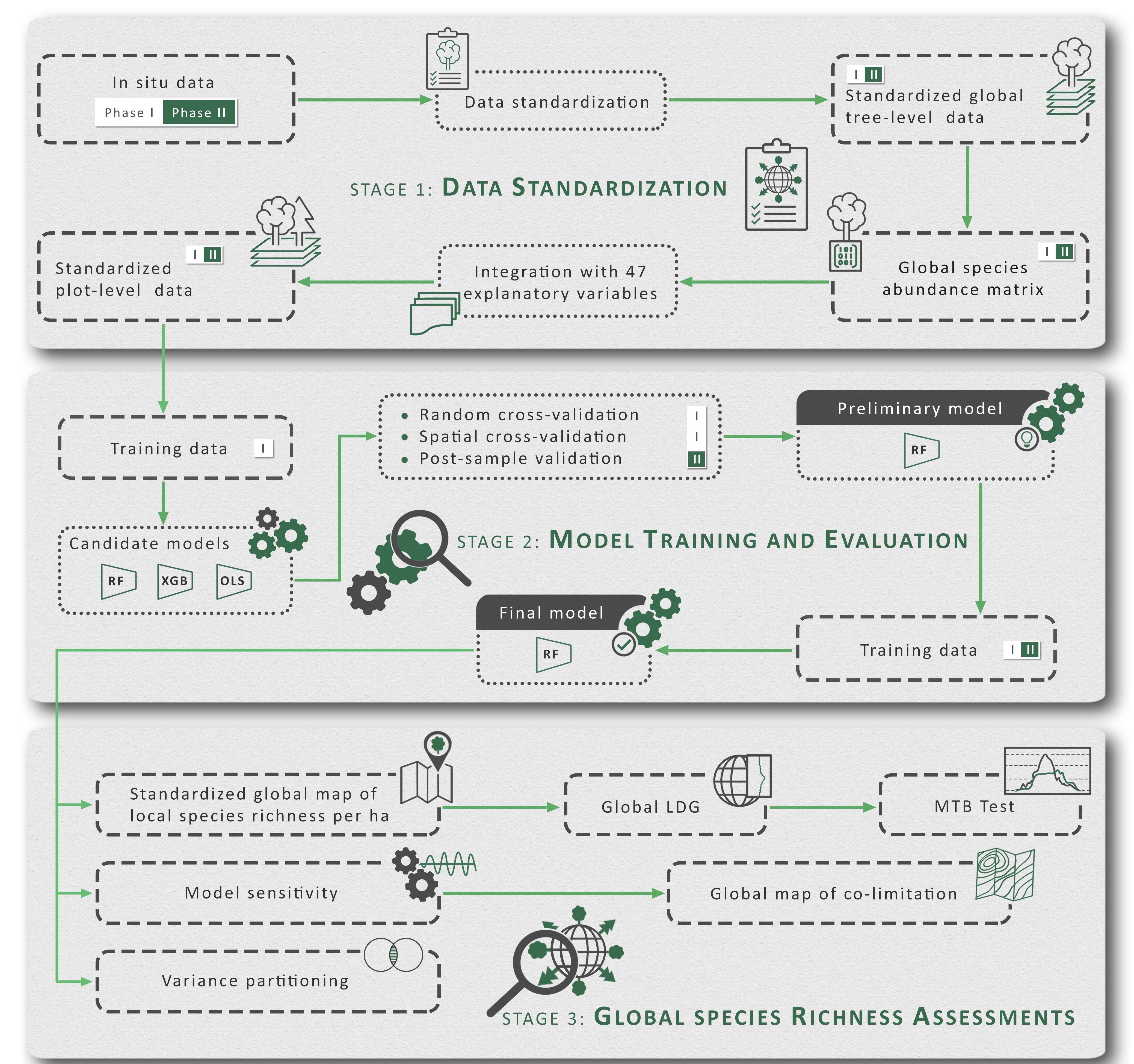
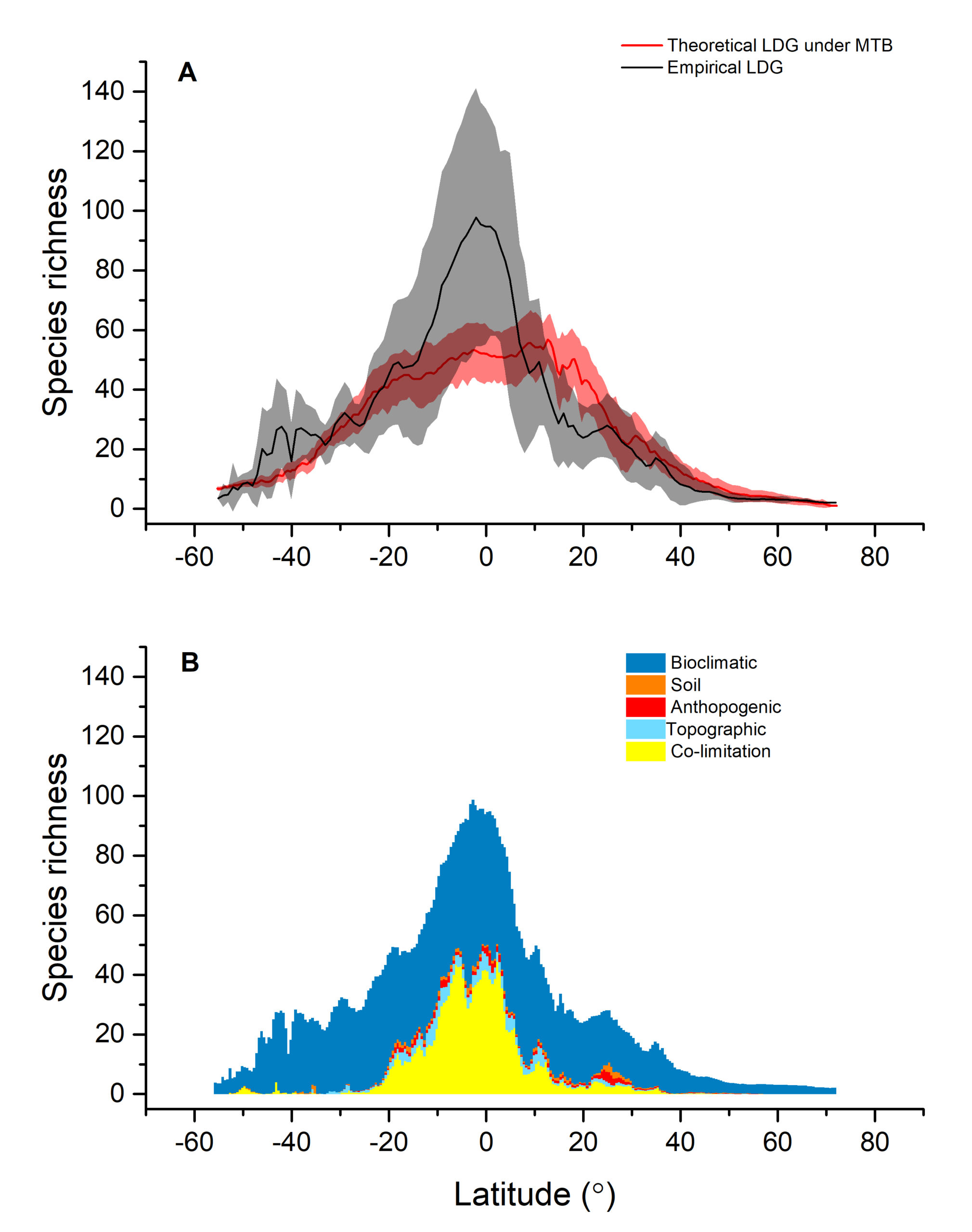
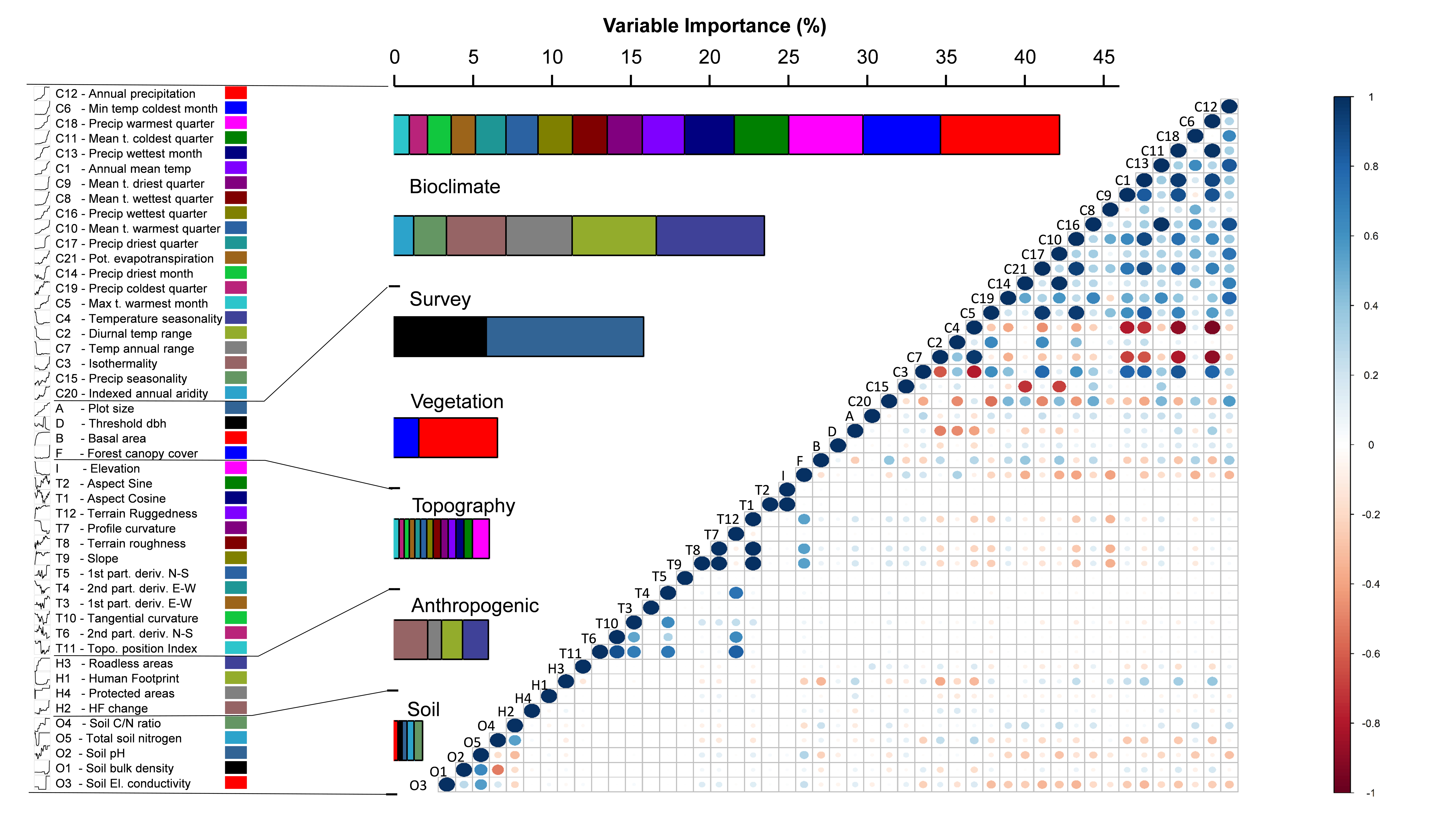
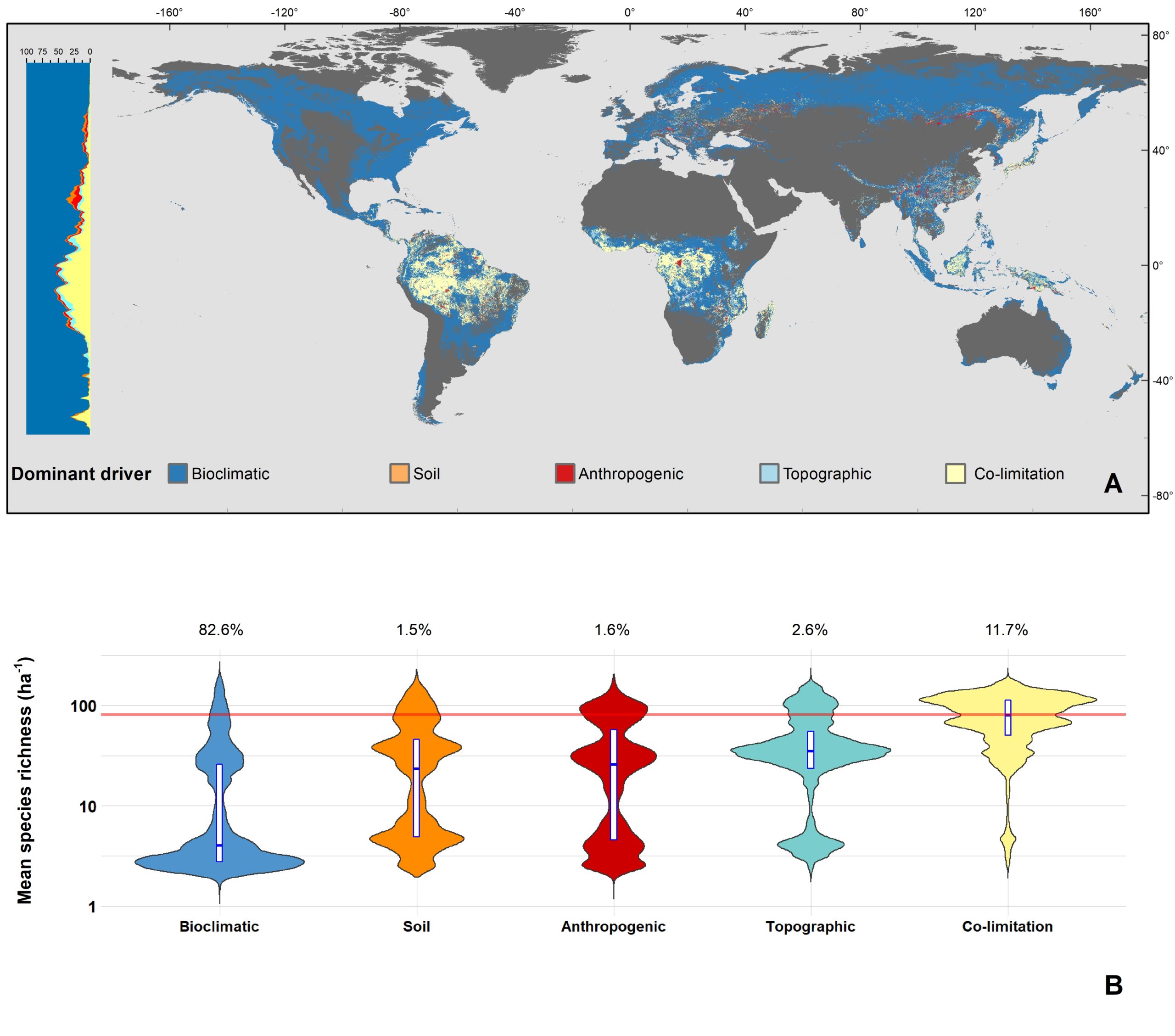
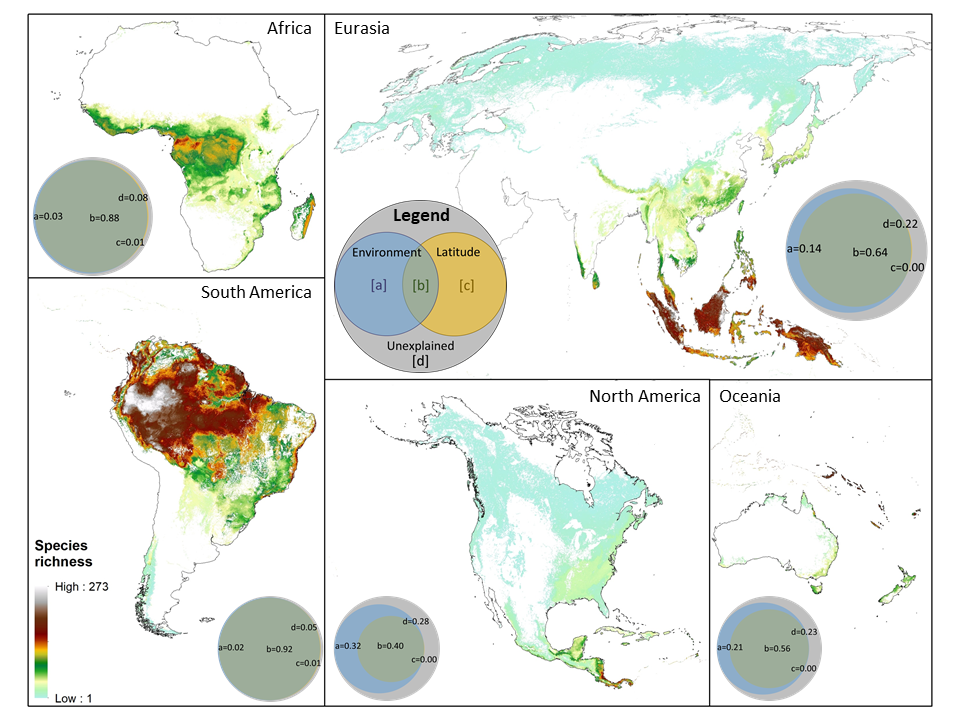
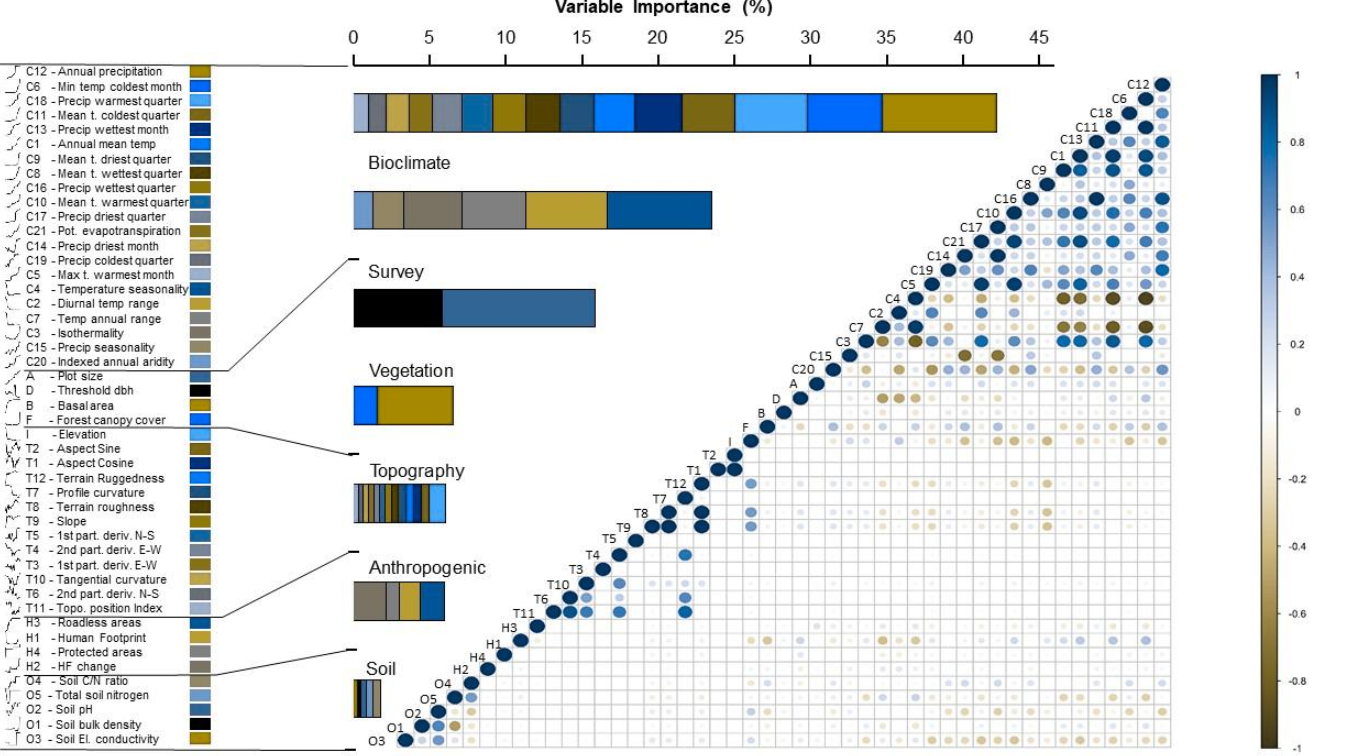
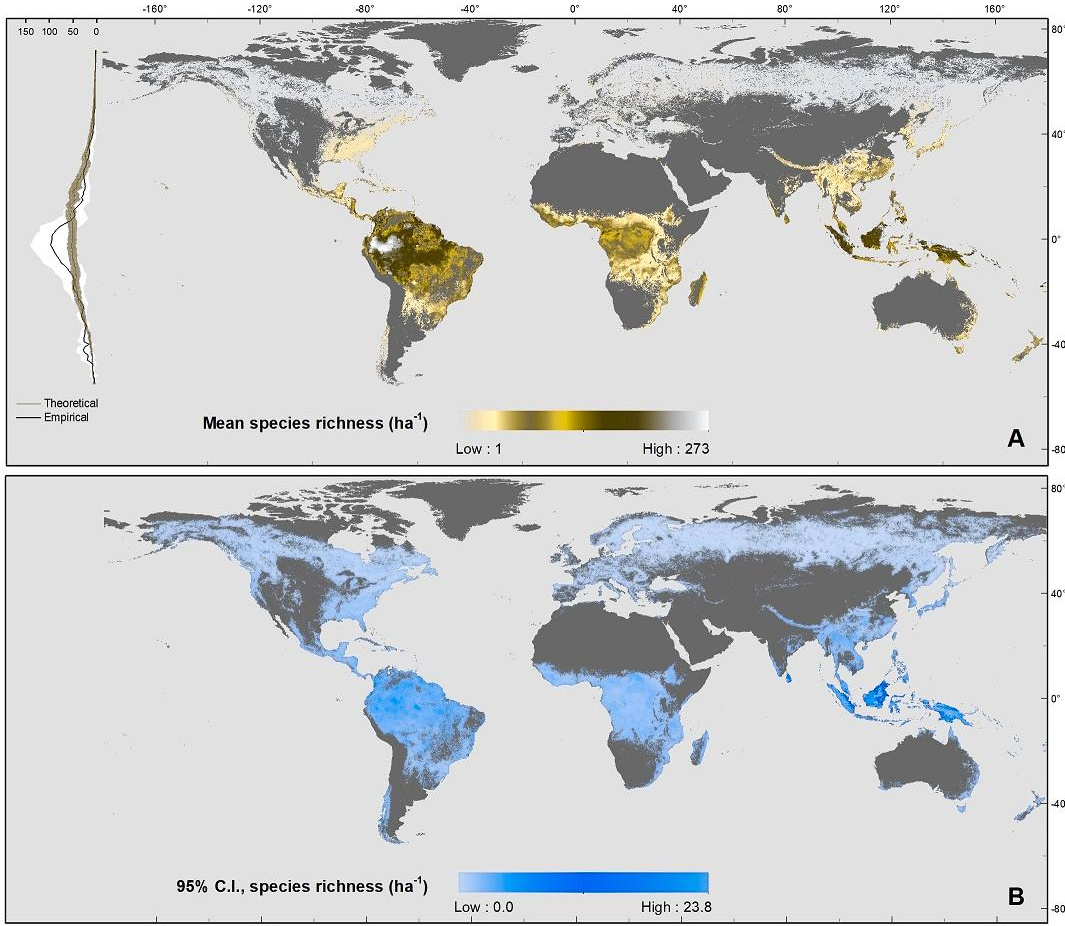
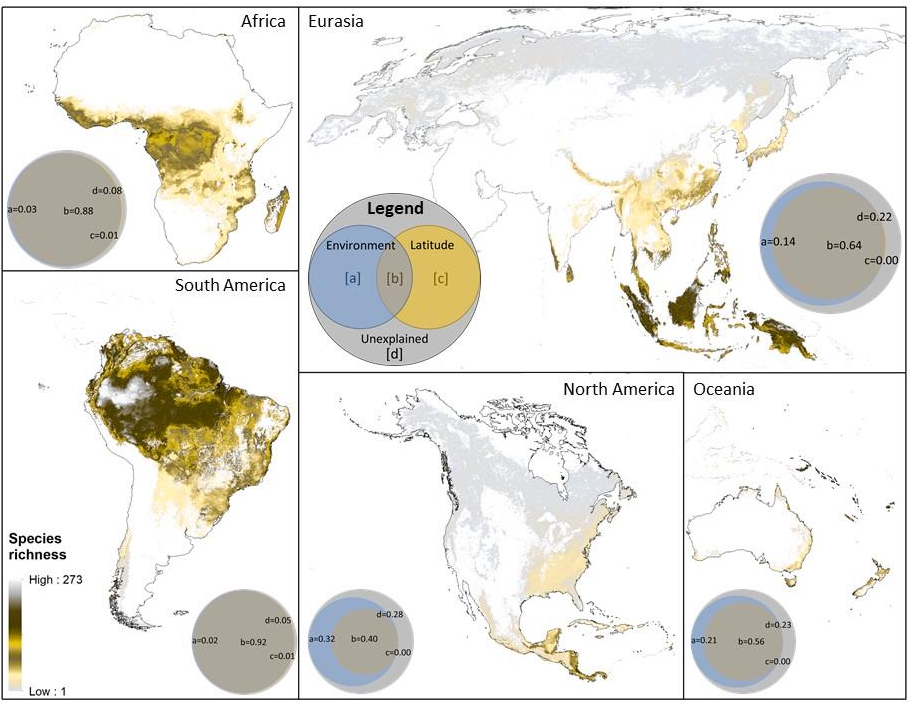
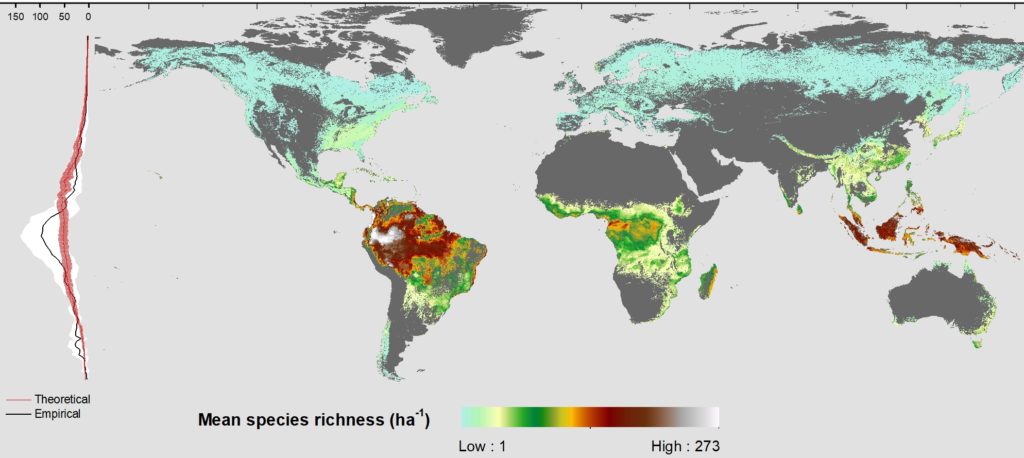
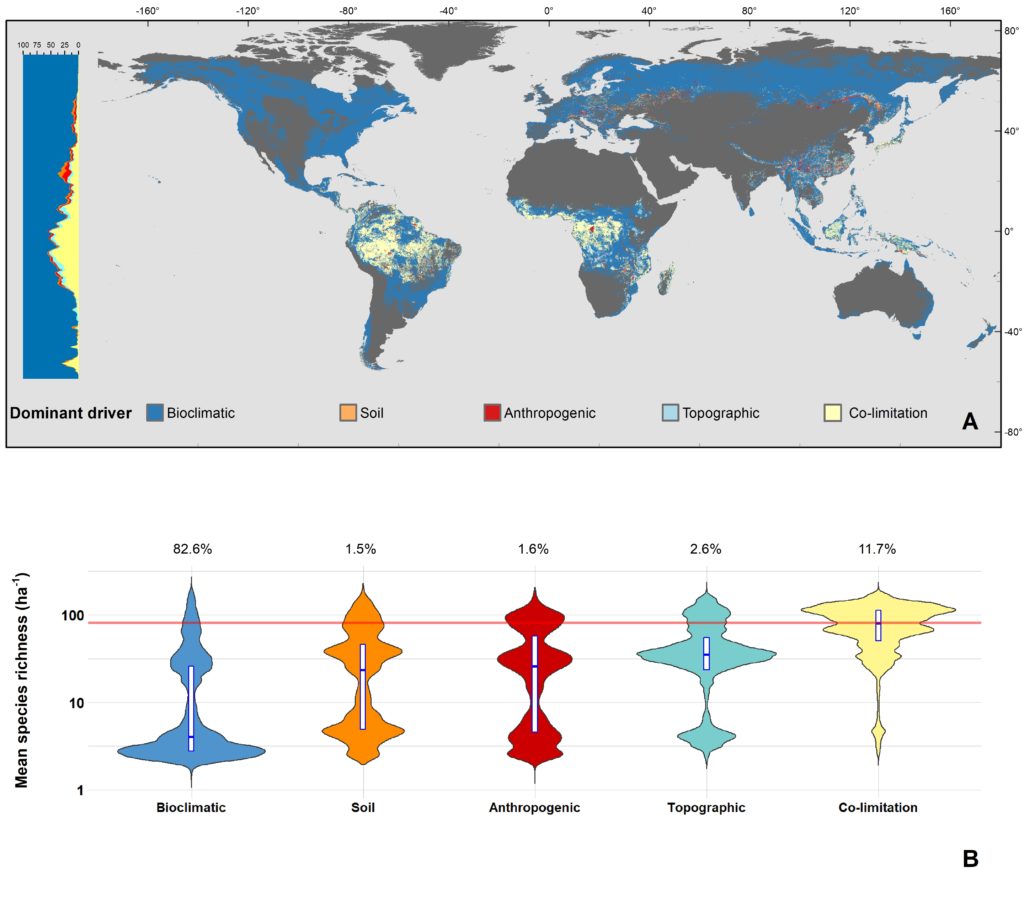
The latitudinal diversity gradient (LDG) is one of the most recognized global patterns of species richness exhibited across a wide range of taxa. Numerous hypotheses have been proposed in the last two centuries to explain LDG, but rigorous tests of the drivers of LDGs have been limited by a lack of high-quality global species richness data. Here, we produce a high-resolution (0.025°×0.025°) map of local tree species richness using a global forest inventory database with individual tree information and local biophysical characteristics from ~1.3 million sample plots. We then quantify drivers of local tree species richness patterns across latitudes. Generally, annual mean temperature was a dominant predictor of tree species richness, which is most consistent with the metabolic theory of biodiversity (MTB). However, MTB underestimated LDG in the tropics, where high species richness was moderated also by topographic, soil, and anthropogenic factors operating at local scales. Given that local landscape variables operate synergistically with bioclimatic factors in shaping the global LDG pattern, we suggest that MTB be extended to account for co-limitation by subordinate drivers.
https://www.nature.com/articles/s41559-022-01831-x
News coverage is currently on the Purdue Agriculture College website as well as the Purdue University website.
Global Map of Local Tree Species Richness per hectare (geoTiff, ~3km resolution)
Global Map of Co-Limitation (geoTiff, ~3km resolution)
Code availability
All the models in this study were constructed using command line applications written in the R programming language, which processed and restructured the input data, trained the model, and performed cross-validation. Due to the massive amount of data, we used Purdue University’s Brown supercomputing cluster to accelerate the training process.
The development of the GFBi database, tabular data cleaning, creation of species abundance matrices, evaluation of diversity determinants, and geostatistical imputation were conducted in R (v.3.4.2) through the use of several Linux-based high-performance computing (HPC) resources at Purdue University, and a custom HPC interface developed
using Amazon Web Services, each designed for batch processing, scalable resource distribution, embarrassingly parallel computations, and/or large RAM jobs. Compute nodes with up to 1TB of RAM and clusters of up to 64 nodes were employed in this study. Portions of the covariate preparation, mapping, and quality control assessment were conducted on Windows-based operating systems with up to 128 GB of RAM.
Final continental-level RF models and the R codes we developed to train the models are available under license MIT, with the identifier: 10.6084/m9.figshare.17234729.
Data availability
- Training dataframe, including Phase-I and Phase-II plot-level data, will be available from the corresponding author (albeca.liang@gmail.com) upon reasonable request and approval from GFBI and data contributors.
- The global map of tree species richness is available under license CC BY 4.0, with the identifier: 10.6084/m9.figshare.17232491;
- The global map of co-limitation is available under license CC BY 4.0, with the identifier: 10.6084/m9.figshare.17234339.
Figures and Downloads
Full Citation
Liang, J., J. G. P. Gamarra, N. Picard, M. Zhou, B. Pijanowski, D. F. Jacobs, P. B. Reich, T. W. Crowther, G.-J. Nabuurs, S. de-Miguel, J. Fang, C. W. Woodall, J.-C. Svenning, T. Jucker, J.-F. Bastin, S. K. Wiser, F. Slik, B. Hérault, G. Alberti, G. Keppel, G. M. Hengeveld, P. L. Ibisch, C. A. Silva, H. ter Steege, P. L. Peri, D. A. Coomes, E. B. Searle, K. von Gadow, B. Jaroszewicz, A. O. Abbasi, M. Abegg, Y. C. A. Yao, J. Aguirre-Gutiérrez, A. M. A. Zambrano, J. Altman, E. Alvarez-Dávila, J. G. Álvarez-González, L. F. Alves, B. H. K. Amani, C. A. Amani, C. Ammer, B. A. Ilondea, C. Antón-Fernández, V. Avitabile, G. A. Aymard, A. F. Azihou, J. A. Baard, T. R. Baker, R. Balazy, M. L. Bastian, R. Batumike, M. Bauters, H. Beeckman, N. M. H. Benu, R. Bitariho, P. Boeckx, J. Bogaert, F. Bongers, O. Bouriaud, P. H. S. Brancalion, S. Brandl, F. Q. Brearley, J. Briseno-Reyes, E. N. Broadbent, H. Bruelheide, E. Bulte, A. C. Catlin, R. Cazzolla Gatti, R. G. César, H. Y. H. Chen, C. Chisholm, E. Cienciala, G. D. Colletta, J. J. Corral-Rivas, A. Cuchietti, A. Cuni-Sanchez, J. A. Dar, S. Dayanandan, T. de Haulleville, M. Decuyper, S. Delabye, G. Derroire, B. DeVries, J. Diisi, T. V. Do, J. Dolezal, A. Dourdain, G. P. Durrheim, N. L. E. Obiang, C. E. N. Ewango, T. J. Eyre, T. M. Fayle, L. F. N. Feunang, L. Finér, M. Fischer, J. Fridman, L. Frizzera, A. L. de Gasper, D. Gianelle, H. B. Glick, M. S. Gonzalez-Elizondo, L. Gorenstein, R. Habonayo, O. J. Hardy, D. J. Harris, A. Hector, A. Hemp, M. Herold, A. Hillers, W. Hubau, T. Ibanez, N. Imai, G. Imani, A. M. Jagodzinski, S. Janecek, V. K. Johannsen, C. A. Joly, B. Jumbam, B. L. P. R. Kabelong, G. A. Kahsay, V. Karminov, K. Kartawinata, J. N. Kassi, E. Kearsley, D. K. Kennard, S. Kepfer-Rojas, M. L. Khan, J. N. Kigomo, H. S. Kim, C. Klauberg, Y. Klomberg, H. Korjus, S. Kothandaraman, F. Kraxner, A. Kumar, R. Kuswandi, M. Lang, M. J. Lawes, R. V. Leite, G. Lentner, S. L. Lewis, M. B. Libalah, J. Lisingo, P. M. López-Serrano, H. Lu, N. V. Lukina, A. M. Lykke, V. Maicher, B. S. Maitner, E. Marcon, A. R. Marshall, E. H. Martin, O. Martynenko, F. M. Mbayu, M. T. E. Mbuvi, J. A. Meave, C. Merow, S. Miscicki, V. S. Moreno, A. Morera, S. A. Mukul, J. C. Müller, A. Murdjoko, M. G. Nava-Miranda, L. E. Ndive, V. J. Neldner, R. V. Nevenic, L. N. Nforbelie, M. L. Ngoh, A. E. N’Guessan, M. R. Ngugi, A. S. K. Ngute, E. N. N. Njila, M. C. Nyako, T. O. Ochuodho, J. Oleksyn, A. Paquette, E. I. Parfenova, M. Park, M. Parren, N. Parthasarathy, S. Pfautsch, O. L. Phillips, M. T. F. Piedade, D. Piotto, M. Pollastrini, L. Poorter, J. R. Poulsen, A. D. Poulsen, H. Pretzsch, M. Rodeghiero, S. G. Rolim, F. Rovero, E. Rutishauser, K. Sagheb-Talebi, P. Saikia, M. N. Sainge, C. Salas-Eljatib, A. Salis, P. Schall, D. Schepaschenko, M. Scherer-Lorenzen, B. Schmid, J. Schöngart, V. Šebeň, G. Sellan, F. Selvi, J. M. Serra-Diaz, D. Sheil, A. Z. Shvidenko, P. Sist, A. F. Souza, K. J. Stereńczak, M. J. P. Sullivan, S. Sundarapandian, M. Svoboda, M. D. Swaine, N. Targhetta, N. Tchebakova, L. A. Trethowan, R. Tropek, J. T. Mukendi, P. M. Umunay, V. A. Usoltsev, G. Vaglio Laurin, R. Valentini, F. Valladares, F. van der Plas, D. J. Vega-Nieva, H. Verbeeck, H. Viana, A. C. Vibrans, S. A. Vieira, J. Vleminckx, C. E. Waite, H.-F. Wang, E. K. Wasingya, C. Wekesa, B. Westerlund, F. Wittmann, V. Wortel, T. Zawiła-Niedźwiecki, C. Zhang, X. Zhao, J. Zhu, X. Zhu, Z.-X. Zhu, I. C. Zo-Bi, and C. Hui. 2022. Co-limitation towards lower latitudes shapes global forest diversity gradients. Nature ecology & evolution 8:10.1038/s41559-41022-01831-x.